Uncover New Insights with Post‑translational Modification (PTM) Analysis
Interrogate PTMs and proteoforms with ease on Platinum® Pro’s next-gen protein sequencing (NGPS) platform, now powered by ProteoVue™ protein variant informatics workflow.
See ProteoVue PaperWhy choose NGPS for PTMs?
Recognize PTM events with unique kinetic signatures
Explore proteoforms at single-molecule resolution
Quantify protein variation at the amino acid level
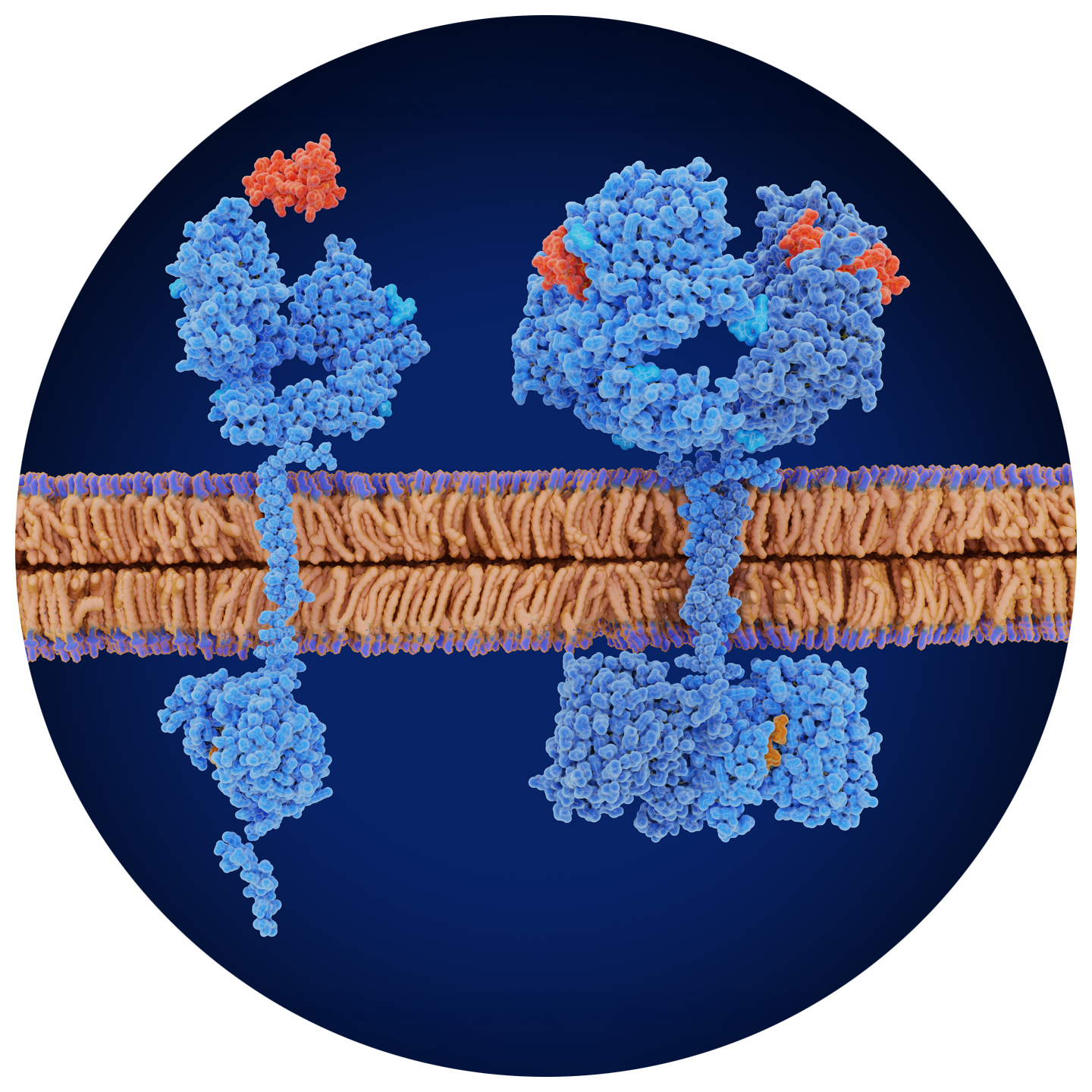
Proteoforms, such as PTMs, play crucial roles in biological and disease mechanisms.
Traditional proteomics techniques such as MS often struggle to capture the full diversity and complexity of proteoforms. NGPS leverages single-molecule sequencing to directly analyze individual protein molecules, providing detailed information on their modi cations and variation. This capability enables previously unattainable methods of protein study, paving the way for new discoveries in biology and medicine.
Generate deeper insights.
Discover more about your proteins. Platinum Pro technology enables you to discover new insights into biological function with the capability to conduct analysis of PTMs. Proteoform research is critical in identifying protein variations associated with phenotypic diversity and understanding complex disease mechanisms. Researchers are actively exploring the human proteome to uncover each protein’s unique function and its contribution to biological processes, disease mechanisms, and cellular interactions.
Quantum-Si invites researchers and potential collaborators to experience the ProteoVue workflow. Join the community of pioneers seeking deeper understanding and groundbreaking discoveries.
See Sequencing SolutionsSee how Platinum Pro’s single-molecule resolution overcomes obstacles presented by other PTM analysis methods:
-
Next-Generation Protein Sequencing and individual ion mass spectrometry enable complementary analysis of IL-6 — PUBLICATION
The vast complexity of the proteome currently overwhelms any single analytical technology in capturing the full spectrum of proteoform diversity. In this study, the team at Northwestern University evaluated the complementarity of two cutting-edge proteomic technologies—single-molecule protein sequencing and individual ion mass spectrometry—for analyzing recombinant human IL-6 (rhIL-6) at the amino acid, peptide, and intact proteoform levels. For single-molecule protein sequencing, they employed the Platinum® instrument. NGPS on Platinum utilizes cycles of N-terminal amino acid recognizer binding and aminopeptidase cleavage to enable parallelized sequencing of single peptide molecules. They found that NGPS produces single amino acid coverage of multiple key regions of IL-6, including two peptides within helices A and C which harbor residues that reportedly impact IL-6 function. For top-down proteoform evaluation, the team used individual ion mass spectrometry (I2MS), a highly parallelized orbitrap-based charge detection MS platform. Single ion detection of gas-phase fragmentation products (I2MS2) gives significant sequence coverage in key regions in IL-6, including two regions within helices B and D that are involved in IL-6 signaling. Together, these complementary technologies delivered a combined 52% sequence coverage, offering a more complete view of IL-6 structural and functional diversity than either technology alone.
Northwestern University’s Neil Kelleher and his team have a published article, featured in Springer’s Analytical and Bioanalytical Chemistry journal. The paper highlights the synergy of complementary protein detection methods to more comprehensively cover protein segments relevant to biological interactions.
-
Detecting Amino Acid Variants Using Next-Generation Protein Sequencing (NGPS) — PREPRINT PUBLICATION
Next-Generation Protein Sequencing (NGPS) is a single-molecule approach for characterizing protein variants, offering detailed insight into proteoforms and amino acid substitutions not easily discerned by mass spectrometry. The novel data type produced by NGPS, which is based on binding of N-terminal amino acids by fluorescently tagged recognizer proteins, requires the development of new data analysis methods and bioinformatic tools.
-
Beyond the Genome: Unraveling Protein Variability with Quantum-Si’s Next-Generation Protein Sequencing Technology – POSTER
Protein sequencing is a groundbreaking advancement in proteomics that augments genomics and transcriptomics research by providing crucial insights into the functional proteins encoded by the genome. Protein sequencing offers a more complete understanding of cellular processes and disease mechanisms by detecting changes at the protein level, such as post-translational modifications (PTMs), which cannot be captured by genomics data alone. NGPS on Platinum enables researchers to identify and characterize proteins with single-molecule resolution in a simple workflow and on a benchtop instrument.